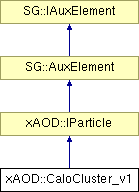
#include <CaloCluster_v1.h>
Public Types | |
| typedef float | flt_t |
| typedef CaloSampling::CaloSample | CaloSample |
Public types | |
| enum | ClusterSize { SW_55ele = 1, SW_35ele = 2, SW_37ele = 3, SW_55gam = 4, SW_35gam = 5, SW_37gam = 6, SW_55Econv = 7, SW_35Econv = 8, SW_37Econv = 9, SW_softe = 10, Topo_420 = 11, Topo_633 = 12, SW_7_11 = 13, CSize_Unknown = 99 } |
Enumeration to identify different cluster sizes. | |
| enum | MomentType { FIRST_PHI = 101, FIRST_ETA = 102, SECOND_R = 201, SECOND_LAMBDA = 202, DELTA_PHI = 301, DELTA_THETA = 302, DELTA_ALPHA = 303, CENTER_X = 401, CENTER_Y = 402, CENTER_Z = 403, CENTER_MAG = 404, CENTER_LAMBDA = 501, LATERAL = 601, LONGITUDINAL = 602, ENG_FRAC_EM = 701, ENG_FRAC_MAX = 702, ENG_FRAC_CORE = 703, FIRST_ENG_DENS = 804, SECOND_ENG_DENS = 805, ISOLATION = 806, ENG_BAD_CELLS = 807, N_BAD_CELLS = 808, N_BAD_CELLS_CORR = 809, BAD_CELLS_CORR_E = 813, BADLARQ_FRAC = 821, ENG_POS = 822, SIGNIFICANCE = 823, CELL_SIGNIFICANCE = 824, CELL_SIG_SAMPLING = 825, AVG_LAR_Q = 826, AVG_TILE_Q = 827, ENG_BAD_HV_CELLS = 828, N_BAD_HV_CELLS = 829, EM_PROBABILITY = 900, HAD_WEIGHT = 901, OOC_WEIGHT = 902, DM_WEIGHT = 903, TILE_CONFIDENCE_LEVEL = 904, VERTEX_FRACTION = 1000, NVERTEX_FRACTION = 1001, ETACALOFRAME = 1100, PHICALOFRAME = 1101, ETA1CALOFRAME = 1102, PHI1CALOFRAME = 1103, ETA2CALOFRAME = 1104, PHI2CALOFRAME = 1105, ENG_CALIB_TOT = 10001, ENG_CALIB_OUT_L = 10010, ENG_CALIB_OUT_M = 10011, ENG_CALIB_OUT_T = 10012, ENG_CALIB_DEAD_L = 10020, ENG_CALIB_DEAD_M = 10021, ENG_CALIB_DEAD_T = 10022, ENG_CALIB_EMB0 = 10030, ENG_CALIB_EME0 = 10031, ENG_CALIB_TILEG3 = 10032, ENG_CALIB_DEAD_TOT = 10040, ENG_CALIB_DEAD_EMB0 = 10041, ENG_CALIB_DEAD_TILE0 = 10042, ENG_CALIB_DEAD_TILEG3 = 10043, ENG_CALIB_DEAD_EME0 = 10044, ENG_CALIB_DEAD_HEC0 = 10045, ENG_CALIB_DEAD_FCAL = 10046, ENG_CALIB_DEAD_LEAKAGE = 10047, ENG_CALIB_DEAD_UNCLASS = 10048, ENG_CALIB_FRAC_EM = 10051, ENG_CALIB_FRAC_HAD = 10052, ENG_CALIB_FRAC_REST = 10053 } |
| enum | State { UNKNOWN = -1, UNCALIBRATED = 0, CALIBRATED = 1, ALTCALIBRATED = 2, NSTATES = 3 } |
enum of possible signal states. | |
Public Member Functions | |
| CaloCluster_v1 () | |
| Default constructor. | |
| CaloCluster_v1 (const CaloCluster_v1 &other) | |
| Copy constructor. | |
| virtual | ~CaloCluster_v1 () |
| Destructor. | |
| CaloCluster_v1 & | operator= (const xAOD::CaloCluster_v1 &other) |
| Assignment operator. | |
| unsigned int | getClusterEtaSize () const |
| Get eta size from cluster size. | |
| unsigned int | getClusterPhiSize () const |
| Get phi size from cluster size. | |
| void | setBadChannelList (const CaloClusterBadChannelList &bcl) |
| const CaloClusterBadChannelList & | badChannelList () const |
| void | toPersistent () |
| Function preparing the object to be persistified. | |
convinience functions | |
| double | et () const |
Energy/Eta/Phi per sampling | |
| float | eSample (const CaloSample sampling) const |
| float | etaSample (const CaloSample sampling) const |
Retrieve barycenter 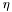 in a given sample. in a given sample. | |
| float | phiSample (const CaloSample sampling) const |
Retrieve barycenter 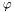 in a given sample. in a given sample. | |
| float | energy_max (const CaloSample sampling) const |
| Retrieve maximum cell energy in given sampling. | |
| float | etamax (const CaloSample sampling) const |
Retrieve 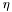 of cell with maximum energy in given sampling. of cell with maximum energy in given sampling. | |
| float | phimax (const CaloSample sampling) const |
Retrieve 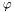 of cell with maximum energy in given sampling. of cell with maximum energy in given sampling. | |
| float | etasize (const CaloSample sampling) const |
Returns cluster size in 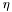 for a given sampling. for a given sampling. | |
| float | phisize (const CaloSample sampling) const |
Returns cluster size in 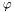 for a given sampling. for a given sampling. | |
| float | energyBE (const unsigned layer) const |
| Get the energy in one layer of the EM Calo. | |
| float | etaBE (const unsigned layer) const |
| Get the eta in one layer of the EM Calo. | |
| float | phiBE (const unsigned layer) const |
| Get the phi in one layer of the EM Calo. | |
| bool | setEnergy (const CaloSample sampling, const float e) |
| Set energy for a given sampling. Returns false if the sample isn't part of the cluster. | |
| bool | setEta (const CaloSample sampling, const float eta) |
Set 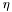 in a given sampling. Returns false if the sample isn't part of the cluster. in a given sampling. Returns false if the sample isn't part of the cluster. | |
| bool | setPhi (const CaloSample sampling, const float phi) |
Set 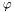 in a given sampling. Returns false if the sample isn't part of the cluster. in a given sampling. Returns false if the sample isn't part of the cluster. | |
| bool | setEmax (const CaloSample sampling, const float eMax) |
| Set the Energy of the cell with the highest energy in a particular sampling. | |
| bool | setEtamax (const CaloSample sampling, const float etaMax) |
| Set the eta of the cell with the highest energy in a particular sampling. | |
| bool | setPhimax (const CaloSample sampling, const float phiMax) |
| Set the phi of the cell with the highest energy in a particular sampling. | |
| bool | setEtasize (const CaloSample sampling, const float etaSize) |
Set the cluster size in 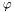 for a given sampling. for a given sampling. | |
| bool | setPhisize (const CaloSample sampling, const float phiSize) |
Set the cluster size in  for a given sampling. for a given sampling. | |
Cluster moments | |
| void | insertMoment (MomentType type, double value) |
| bool | retrieveMoment (MomentType type, double &value) const |
| Retrieve individual moment. | |
| double | getMomentValue (MomentType type) const |
| Retrieve individual moment - no check for existance! Returns -999 on error. | |
Special Cluster Variables | |
| void | setEta0 (flt_t) |
| flt_t | eta0 () const |
Returns raw 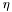 of cluster seed. of cluster seed. | |
| void | setPhi0 (flt_t) |
Set raw  of cluster seed. of cluster seed. | |
| flt_t | phi0 () const |
Returns raw  of cluster seed. of cluster seed. | |
| void | setTime (flt_t) |
| Set cluster time. | |
| flt_t | time () const |
| Access cluster time. | |
| unsigned | samplingPattern () const |
| Access to sampling pattern (one bit per sampling) (Method may be removed later). | |
| void | setSamplingPattern (const unsigned sp, const bool clearSamplingVars=false) |
| Set sampling pattern (one bit per sampling. | |
| void | clearSamplingData () |
| Clear the sampling data. | |
| unsigned | nSamples () const |
| bool | hasSampling (const CaloSample s) const |
| Checks if certain smapling contributes to cluster. | |
| ClusterSize | clusterSize () const |
| Get cluster size. | |
| void | setClusterSize (const ClusterSize) |
| Get cluster size. | |
| bool | inBarrel () const |
| Returns true if at least one clustered cell in the barrel. | |
| bool | inEndcap () const |
| Returns true if at least one clustered cell in the endcap. | |
4Momentum setter (signal-state dependent) | |
| void | setE (flt_t) |
| void | setEta (flt_t) |
| Set Eta for the current signal state. | |
| void | setPhi (flt_t) |
| Set Phi for the current signal state. | |
| void | setM (flt_t) |
| Set Mass for the current signal state. | |
Friends | |
| class | ::CaloClusterChangeSignalState |
Functions implementing the xAOD::IParticle interface | |
|
| |
| typedef IParticle::FourMom_t | FourMom_t |
| Definition of the 4-momentum type. | |
| virtual double | pt () const |
The transverse momentum (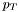 ) of the particle (negative for negative-energy clusters). ) of the particle (negative for negative-energy clusters). | |
| virtual double | eta () const |
The pseudorapidity (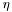 ) of the particle. ) of the particle. | |
| virtual double | phi () const |
The azimuthal angle (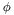 ) of the particle. ) of the particle. | |
| virtual double | m () const |
| The invariant mass of the particle. | |
| virtual double | e () const |
| The total energy of the particle. | |
| virtual double | rapidity () const |
| The true rapidity (y) of the particle. | |
| virtual const FourMom_t & | p4 () const |
| The full 4-momentum of the particle. | |
| const FourMom_t & | p4 (const State s) const |
| virtual Type::ObjectType | type () const |
| The type of the object as a simple enumeration. | |
Athena-only methods, used during building stage | |
Set up an ElementLink to a CaloClusterCellLink object | |
|
typedef CaloClusterCellLink::const_iterator | const_cell_iterator |
| Iterator of the underlying CaloClusterCellLink (const version). | |
|
typedef CaloClusterCellLink::iterator | cell_iterator |
| Iterator of the underlying CaloClusterCellLink (non-const version). | |
| typedef const_cell_iterator | const_iterator |
| STL-compatible iterators. | |
| typedef cell_iterator | iterator |
| void | addCellLink (CaloClusterCellLink *CCCL) |
| bool | setLink (CaloClusterCellLinkContainer *CCCL, IProxyDictWithPool *sg=nullptr) |
| Set up an ElementLink to a CaloClusterCellLink object. | |
| const CaloClusterCellLink * | getCellLinks () const |
| Get a pointer to the CaloClusterCellLink object (const version). | |
| CaloClusterCellLink * | getCellLinks () |
| Get a pointer to the CaloClusterCellLink object (const version). | |
| bool | addCell (const unsigned index, const double weight) |
| Method to add a cell to the cluster (Beware: Kinematics not updated!). | |
| bool | removeCell (const CaloCell *ptr) |
| Method to remove a cell to the cluster (slow!) (Beware: Kinematics not updated!). | |
| size_t | size () const |
| size method (forwarded from CaloClusterCellLink obj) | |
| const_cell_iterator | cell_begin () const |
| const_cell_iterator | cell_end () const |
| cell_iterator | cell_begin () |
| cell_iterator | cell_end () |
| const_iterator | begin () const |
| const_iterator | end () const |
| iterator | begin () |
| iterator | end () |
| void | reweightCell (cell_iterator it, const double weight) |
| Method to reweight a cell in the cluster (Beware: Kinematics not updated!). | |
| CaloRecoStatus & | recoStatus () |
| Accesssor to CaloRecoStatus (non-const). | |
| const CaloRecoStatus & | recoStatus () const |
| Accesssor to CaloRecoStatus (const). | |
4Momentum in different signal states | |
|
Get Energy in signal state UNCALIBRATED | |
| flt_t | rawE () const |
| void | setRawE (flt_t) |
| Set Energy for signal state UNCALIBRATED. | |
| flt_t | rawEta () const |
Get 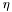 in signal state UNCALIBRATED. in signal state UNCALIBRATED. | |
| void | setRawEta (flt_t) |
Set 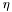 for signal state UNCALIBRATED. for signal state UNCALIBRATED. | |
| flt_t | rawPhi () const |
Get  in signal state UNCALIBRATED. in signal state UNCALIBRATED. | |
| void | setRawPhi (flt_t) |
Set  for signal state UNCALIBRATED. for signal state UNCALIBRATED. | |
| flt_t | rawM () const |
| Get mass in signal state UNCALIBRATED. | |
| void | setRawM (flt_t) |
| Set mass for singal state UNCALIBRATED. | |
| flt_t | altE () const |
| Get Energy in signal state ALTCALIBRATED. | |
| void | setAltE (flt_t) |
| Set Energy for signal state ALTCALIBRATED. | |
| flt_t | altEta () const |
Get 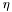 in signal state ALTCALIBRATED. in signal state ALTCALIBRATED. | |
| void | setAltEta (flt_t) |
Set 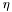 for signal state ALTCALIBRATED. for signal state ALTCALIBRATED. | |
| flt_t | altPhi () const |
Get  in signal state ALTCALIBRATED. in signal state ALTCALIBRATED. | |
| void | setAltPhi (flt_t) |
Set  for signal state ALTCALIBRATED. for signal state ALTCALIBRATED. | |
| flt_t | altM () const |
| Get mass in signal state ALTCALIBRATED. | |
| void | setAltM (flt_t) |
| Set mass for singal state ALTCALIBRATED. | |
| flt_t | calE () const |
| Geet Energy in signal state CALIBRATED. | |
| void | setCalE (flt_t) |
| Set Energy for signal state CALIBRATED. | |
| flt_t | calEta () const |
Get 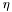 in signal state CALIBRATED. in signal state CALIBRATED. | |
| void | setCalEta (flt_t) |
Set 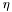 for signal state CALIBRATED. for signal state CALIBRATED. | |
| flt_t | calPhi () const |
Get  in signal state CALIBRATED. in signal state CALIBRATED. | |
| void | setCalPhi (flt_t) |
Set  for signal state CALIBRATED. for signal state CALIBRATED. | |
| flt_t | calM () const |
| Get mass in signal state CALIBRATED. | |
| void | setCalM (flt_t) |
| Set mass for singal state CALIBRATED. | |
| State | signalState () const |
| Get the current signal state. | |
| double | pt (const State s) const |
| pt with a given signal state | |
| double | e (const State s) const |
| energy with a given signal state | |
| double | eta (const State s) const |
| eta with a given signal state | |
| double | phi (const State s) const |
| eta with a given signal state | |
Description of a calorimeter cluster
Enums to identify different moments
Each enum corresponds to a different characteristic moment of a CaloCluster. The enums are loosly grouped by their type - i.e. first moment over an angle, second moment, angular deviations etc.
| bool xAOD::CaloCluster_v1::addCell | ( | const unsigned | index, | |
| const double | weight | |||
| ) | [inline] |
Method to add a cell to the cluster (Beware: Kinematics not updated!).
| index | Index of the cell in the CaloCelLContainer | |
| weight | Weight of the cell in the cluster |
| double xAOD::CaloCluster_v1::e | ( | const State | s | ) | const |
energy with a given signal state
Return eta for a specific signal state.
| s | The desired signal state. |
| float xAOD::CaloCluster_v1::energyBE | ( | const unsigned | layer | ) | const |
Get the energy in one layer of the EM Calo.
| layer | Layer between 0 (Presampler) and 3 (Back) |
| double xAOD::CaloCluster_v1::eta | ( | const State | s | ) | const |
eta with a given signal state
Return eta for a specific signal state.
| s | The desired signal state. |
| float xAOD::CaloCluster_v1::etaBE | ( | const unsigned | layer | ) | const |
Get the eta in one layer of the EM Calo.
| layer | Layer between 0 (Presampler) and 3 (Back) |
| CaloClusterCellLink* xAOD::CaloCluster_v1::getCellLinks | ( | ) | [inline] |
Get a pointer to the CaloClusterCellLink object (const version).
| const CaloClusterCellLink * xAOD::CaloCluster_v1::getCellLinks | ( | ) | const |
Get a pointer to the CaloClusterCellLink object (const version).
| unsigned int xAOD::CaloCluster_v1::getClusterEtaSize | ( | ) | const |
Get eta size from cluster size.
for debugging only ... std::vector<std::pair<std::string,float> > CaloCluster_v1::getAllMoments() { std::vector<std::pair<std::string,float> > retval; const SG::auxid_set_t& auxIds=container()->getAuxIDs(); //->getDynamicAuxIDs(); const size_t idx= this->index(); for (auto ai: auxIds) { const std::string& auxName=SGAuxTypeRegistry::instance().getName(ai); const float v=container()->getData<float>(ai,idx); std::cout << "Index=" <<idx << ", Auxid=" << ai << ", Name=" << auxName << " value=" << v << std::endl; retval.push_back(std::make_pair(auxName,v)); } return retval; }
| double xAOD::CaloCluster_v1::phi | ( | const State | s | ) | const |
eta with a given signal state
Return phi for a specific signal state.
| s | The desired signal state. |
| float xAOD::CaloCluster_v1::phiBE | ( | const unsigned | layer | ) | const |
Get the phi in one layer of the EM Calo.
| layer | Layer between 0 (Presampler) and 3 (Back) |
| double xAOD::CaloCluster_v1::pt | ( | const State | s | ) | const |
pt with a given signal state
Notice that this function is very slow for calorimeter clusters, so it should be called as few times as possible.
| const CaloRecoStatus& xAOD::CaloCluster_v1::recoStatus | ( | ) | const [inline] |
Accesssor to CaloRecoStatus (const).
| CaloRecoStatus& xAOD::CaloCluster_v1::recoStatus | ( | ) | [inline] |
Accesssor to CaloRecoStatus (non-const).
| bool xAOD::CaloCluster_v1::removeCell | ( | const CaloCell * | ptr | ) |
Method to remove a cell to the cluster (slow!) (Beware: Kinematics not updated!).
| index | Index of the cell in the CaloCelLContainer |
| void xAOD::CaloCluster_v1::reweightCell | ( | cell_iterator | it, | |
| const double | weight | |||
| ) | [inline] |
Method to reweight a cell in the cluster (Beware: Kinematics not updated!).
| it | Non-const cell-iterator | |
| weight | The new weight |
| bool xAOD::CaloCluster_v1::setLink | ( | CaloClusterCellLinkContainer * | CCCL, | |
| IProxyDictWithPool * | sg = nullptr | |||
| ) |
Set up an ElementLink to a CaloClusterCellLink object.
| CCCL_key | StoreGate key of the CaloClusterCellLinkContainer | |
| index | Index of inside the CaloClusterCellLinkContainer container |
| CCCL | pointer to the CaloClusterCellLinkContainer | |
| sg | Explicitly specify the store to use for the ElementLink. |
| size_t xAOD::CaloCluster_v1::size | ( | ) | const [inline] |
size method (forwarded from CaloClusterCellLink obj)
| void xAOD::CaloCluster_v1::toPersistent | ( | ) |
Function preparing the object to be persistified.
This function takes care of preparing (all) the ElementLink(s) in the object to be persistified.
 1.6.1
1.6.1